library(tidymodels)
library(tidyverse)
library(here)
library(dotwhisker)Model Evaluation
Library packages
Load Data
symptoms_fit <- readRDS(here("fluanalysis/data/processed_data/symptoms_clean.RDS"))
colnames(symptoms_fit) #quick look at data [1] "SwollenLymphNodes" "ChestCongestion" "ChillsSweats"
[4] "NasalCongestion" "CoughYN" "Sneeze"
[7] "Fatigue" "SubjectiveFever" "Headache"
[10] "Weakness" "WeaknessYN" "CoughIntensity"
[13] "CoughYN2" "Myalgia" "MyalgiaYN"
[16] "RunnyNose" "AbPain" "ChestPain"
[19] "Diarrhea" "EyePn" "Insomnia"
[22] "ItchyEye" "Nausea" "EarPn"
[25] "Hearing" "Pharyngitis" "Breathless"
[28] "ToothPn" "Vision" "Vomit"
[31] "Wheeze" "BodyTemp" Data Split
data_split <- initial_split(symptoms_fit, prop = 3/4) # 75% of data goes into training set
train_data <- training(data_split)
test_data <- testing(data_split)Model 1 fitting: all variables to predict nausea
Define the model: logistic regression
log_mod <- logistic_reg() %>% #model type is logistic regression
set_engine("glm") #engine set to generalized linear modelI am using a logistic regression here because the outcome of interest (Nausea Y/N) is categorical
Create recipe
recipe_nausea <- recipe(Nausea ~., data = train_data)Nausea is the outcome and all other variables are predictors
Create workflow: combine model definition and recipe
nausea_log_wf <- workflow() %>%
add_model(log_mod) %>% #model definition
add_recipe(recipe_nausea) #model recipeThis will run a logistic regression on the flu data, predicting nausea using all other variables that we kept.
Model fitting
set.seed(626)
nausea_fit <- nausea_log_wf %>%
fit(data = train_data)
nausea_fit %>% extract_fit_parsnip() %>%
tidy()# A tibble: 38 × 5
term estimate std.error statistic p.value
<chr> <dbl> <dbl> <dbl> <dbl>
1 (Intercept) 0.386 9.31 0.0414 0.967
2 SwollenLymphNodesYes -0.0795 0.230 -0.346 0.729
3 ChestCongestionYes 0.110 0.249 0.440 0.660
4 ChillsSweatsYes 0.743 0.369 2.02 0.0438
5 NasalCongestionYes 0.344 0.300 1.14 0.252
6 CoughYNYes -0.206 0.613 -0.336 0.737
7 SneezeYes 0.217 0.254 0.855 0.393
8 FatigueYes 0.157 0.423 0.371 0.711
9 SubjectiveFeverYes -0.157 0.272 -0.579 0.563
10 HeadacheYes 0.272 0.333 0.814 0.415
# … with 28 more rowsModel assessment on training data: ROC curve
set.seed(626)
nausea_aug_test <- augment(nausea_fit, train_data)
nausea_aug_test %>%
roc_curve(truth = Nausea, .pred_Yes, event_level = "second") %>%
autoplot()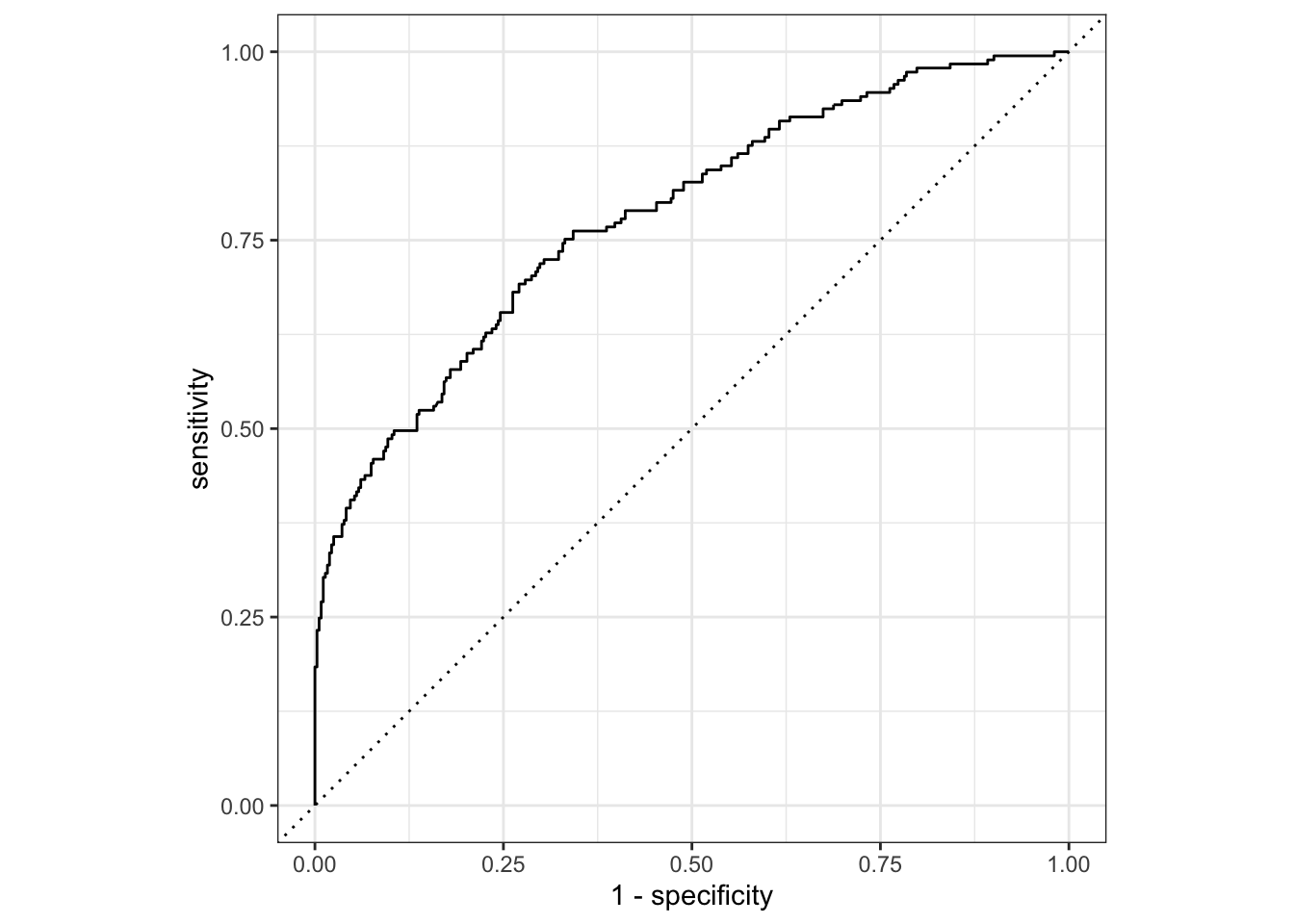
nausea_aug_test %>% roc_auc(truth = Nausea, .pred_Yes,
event_level = "second")# A tibble: 1 × 3
.metric .estimator .estimate
<chr> <chr> <dbl>
1 roc_auc binary 0.782ROC-AUC is ok
Model assessment on testing data: ROC curve
set.seed(626)
nausea_fit_test <- nausea_log_wf %>%
fit(data = test_data)
nausea_fit_test %>% extract_fit_parsnip() %>%
tidy()# A tibble: 38 × 5
term estimate std.error statistic p.value
<chr> <dbl> <dbl> <dbl> <dbl>
1 (Intercept) -10.6 18.7 -0.566 0.571
2 SwollenLymphNodesYes -1.07 0.481 -2.22 0.0263
3 ChestCongestionYes 0.779 0.504 1.55 0.122
4 ChillsSweatsYes -0.685 0.608 -1.13 0.260
5 NasalCongestionYes 0.592 0.573 1.03 0.301
6 CoughYNYes -0.310 1.05 -0.294 0.769
7 SneezeYes 0.478 0.458 1.04 0.297
8 FatigueYes 0.223 1.08 0.208 0.835
9 SubjectiveFeverYes 1.72 0.556 3.10 0.00195
10 HeadacheYes 0.542 0.688 0.788 0.430
# … with 28 more rowsnausea_aug_test2 <- augment(nausea_fit_test, test_data)
nausea_aug_test2 %>%
roc_curve(truth = Nausea, .pred_Yes, event_level = "second") %>%
autoplot()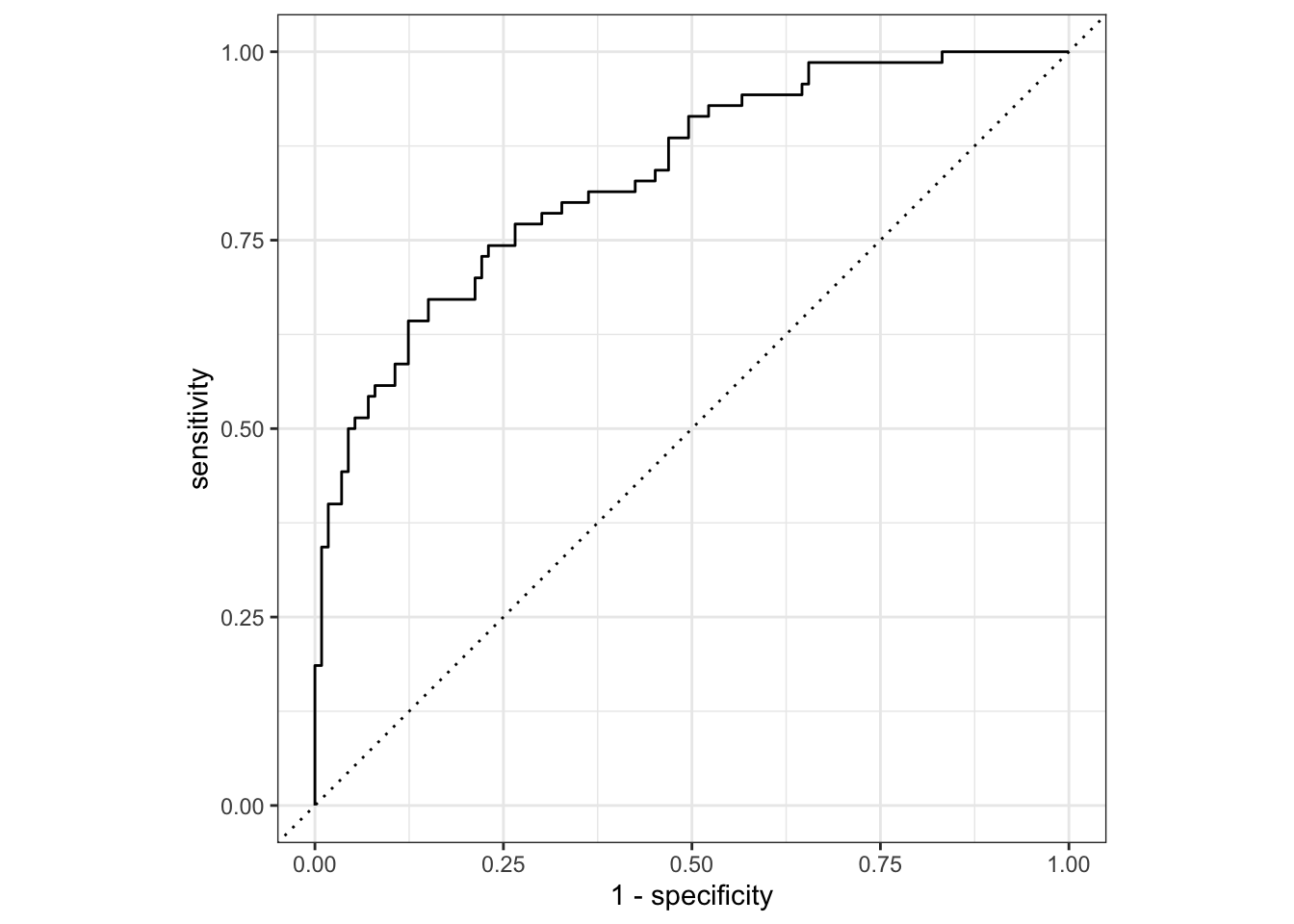
nausea_aug_test2 %>% roc_auc(truth = Nausea, .pred_Yes,
event_level = "second")# A tibble: 1 × 3
.metric .estimator .estimate
<chr> <chr> <dbl>
1 roc_auc binary 0.836The testing data out-performed the training set
Model 2 fitting: runny nose to predict nausea
Create new recipe and workflow
recipe_nausea2 <- recipe(Nausea ~RunnyNose, data = train_data) #only include runny nose
nausea_log_wf2 <- workflow() %>%
add_model(log_mod) %>% #model definition, use the same as Model 1
add_recipe(recipe_nausea2) #model recipeModel fitting
set.seed(626)
nausea_fit2 <- nausea_log_wf2 %>%
fit(data = train_data)
nausea_fit2 %>% extract_fit_parsnip() %>%
tidy()# A tibble: 2 × 5
term estimate std.error statistic p.value
<chr> <dbl> <dbl> <dbl> <dbl>
1 (Intercept) -0.738 0.165 -4.46 0.00000806
2 RunnyNoseYes 0.0961 0.198 0.487 0.626 Model assessment on training data
set.seed(626)
nausea_aug_test2 <- augment(nausea_fit2, train_data)
nausea_aug_test2 %>%
roc_curve(truth = Nausea, .pred_Yes, event_level = "second") %>%
autoplot()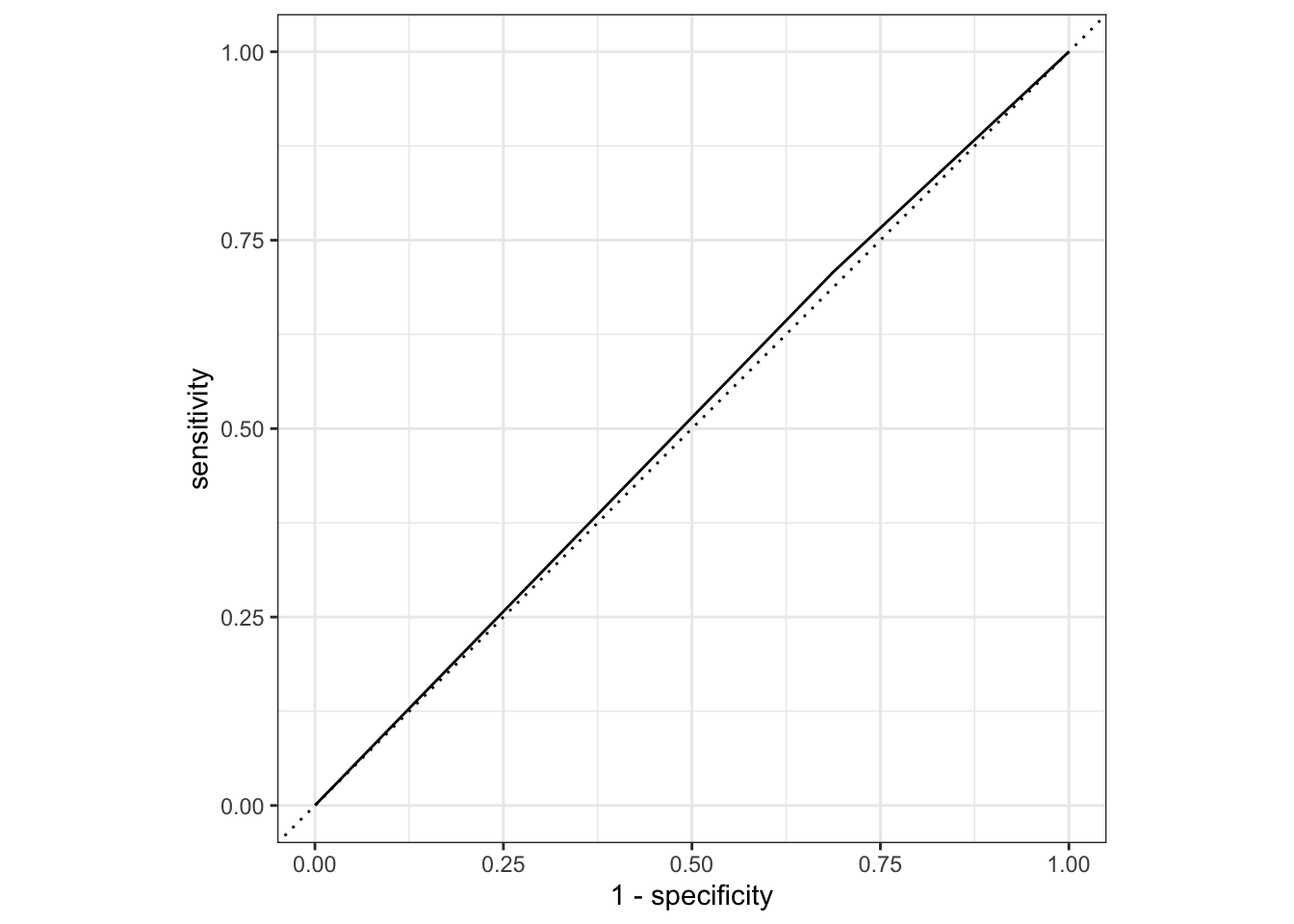
nausea_aug_test2 %>% roc_auc(truth = Nausea, .pred_Yes,
event_level = "second")# A tibble: 1 × 3
.metric .estimator .estimate
<chr> <chr> <dbl>
1 roc_auc binary 0.510The ROC-AUC here is lower than Model 1
Model assessment on testing data
set.seed(626)
nausea_fit_test3 <- nausea_log_wf2 %>%
fit(data = test_data)
nausea_fit_test3 %>% extract_fit_parsnip() %>%
tidy()# A tibble: 2 × 5
term estimate std.error statistic p.value
<chr> <dbl> <dbl> <dbl> <dbl>
1 (Intercept) -0.368 0.307 -1.20 0.230
2 RunnyNoseYes -0.147 0.353 -0.416 0.677nausea_aug_test3 <- augment(nausea_fit_test3, test_data)
nausea_aug_test3 %>%
roc_curve(truth = Nausea, .pred_Yes, event_level = "second") %>%
autoplot()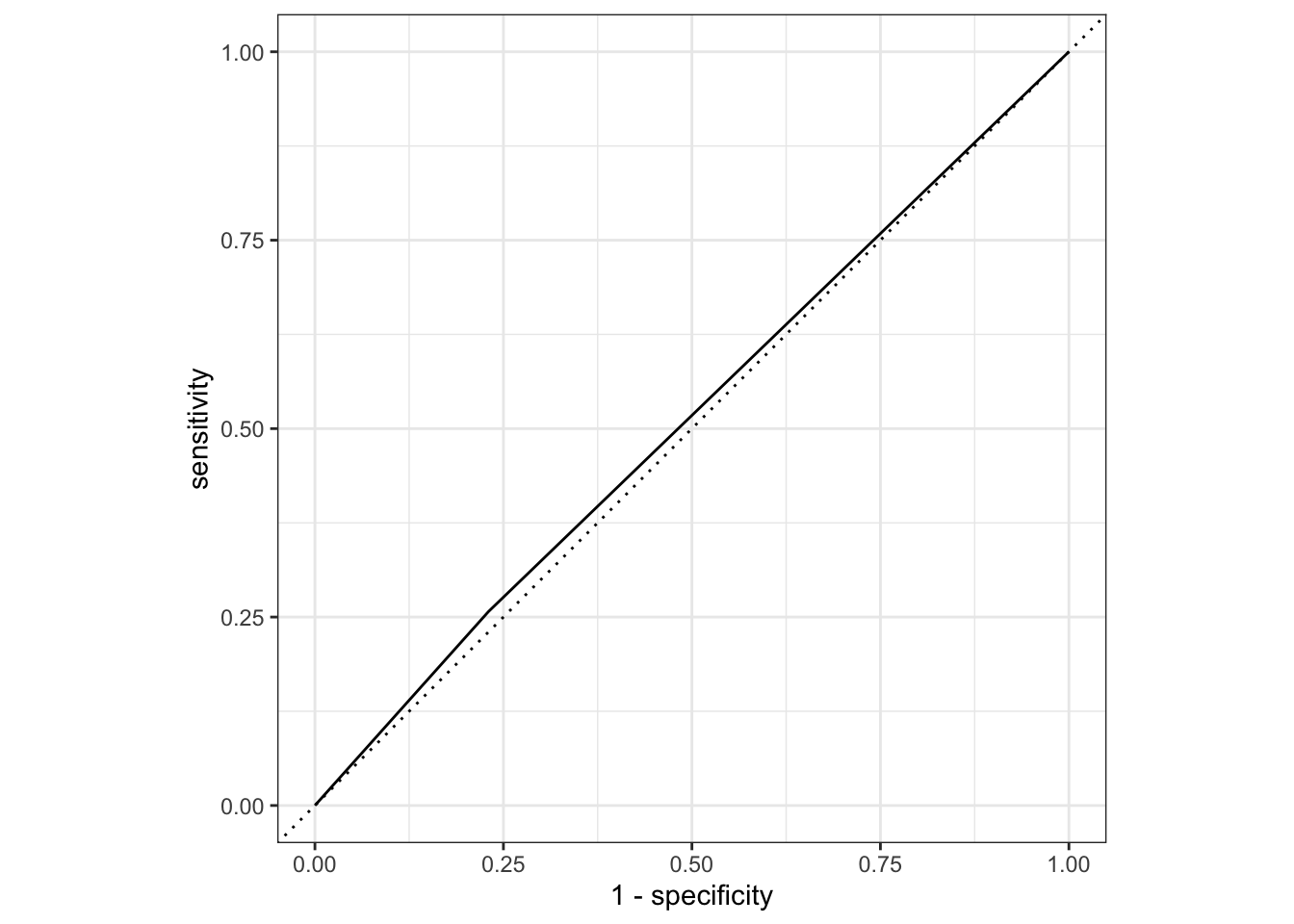
nausea_aug_test3 %>% roc_auc(truth = Nausea, .pred_Yes,
event_level = "second")# A tibble: 1 × 3
.metric .estimator .estimate
<chr> <chr> <dbl>
1 roc_auc binary 0.514The testing data is no different. Runny nose is likely not a good predictor of nausea.
This Section added by Nathan Greenslit
Model 3 fitting: all variables to predict Body Temperature
Define the model: linear regression
lin_mod <- linear_reg() %>% #model type is linear regression
set_engine("lm") #engine set to linear modelCreate recipe for Multivariate model
recipe_bt_all <- recipe(BodyTemp ~., data = train_data)Body Temp is the outcome and all other variables are predictors
Create workflow: combine model definition and recipe
bt_lin_wf_all <- workflow() %>%
add_model(lin_mod) %>% #model definition
add_recipe(recipe_bt_all) #model recipeThis will run a linear regression on the flu data, predicting body temperature using all other variables that we kept.
Multivariate Model Fitting
bt_fit_all <- bt_lin_wf_all %>%
fit(data = train_data)
bt_fit_all %>% extract_fit_parsnip() %>%
tidy()# A tibble: 38 × 5
term estimate std.error statistic p.value
<chr> <dbl> <dbl> <dbl> <dbl>
1 (Intercept) 97.9 0.354 277. 0
2 SwollenLymphNodesYes -0.204 0.108 -1.89 0.0599
3 ChestCongestionYes 0.0863 0.114 0.759 0.448
4 ChillsSweatsYes 0.100 0.158 0.637 0.525
5 NasalCongestionYes -0.376 0.134 -2.81 0.00517
6 CoughYNYes 0.459 0.291 1.58 0.115
7 SneezeYes -0.470 0.117 -4.02 0.0000658
8 FatigueYes 0.424 0.181 2.35 0.0191
9 SubjectiveFeverYes 0.488 0.125 3.89 0.000112
10 HeadacheYes 0.0343 0.146 0.235 0.815
# … with 28 more rowsModel assessment on training data: RMSE and Rsq
set.seed(626)
bt_aug_train_all <- augment(bt_fit_all, train_data) #pull in train dataWarning in predict.lm(object = object$fit, newdata = new_data, type =
"response"): prediction from a rank-deficient fit may be misleading### RMSE and R2 to test model performance
rmse1 <- bt_aug_train_all %>% rmse(truth = BodyTemp, .pred) #RMSE
rsq1 <- bt_aug_train_all %>% rsq(truth = BodyTemp, .pred) #RSQ
bt_metrics_all_train<- full_join(rmse1, rsq1)Joining with `by = join_by(.metric, .estimator, .estimate)`bt_metrics_all_train# A tibble: 2 × 3
.metric .estimator .estimate
<chr> <chr> <dbl>
1 rmse standard 1.12
2 rsq standard 0.166Model assessment on testing data: RMSE and Rsq
set.seed(626)
bt_fit_test_all <- bt_lin_wf_all %>%
fit(data = test_data) #pull in test data
bt_fit_test_all %>% extract_fit_parsnip() %>%
tidy()# A tibble: 38 × 5
term estimate std.error statistic p.value
<chr> <dbl> <dbl> <dbl> <dbl>
1 (Intercept) 98.7 0.655 151. 6.80e-164
2 SwollenLymphNodesYes -0.0195 0.186 -0.105 9.17e- 1
3 ChestCongestionYes 0.297 0.205 1.45 1.49e- 1
4 ChillsSweatsYes 0.279 0.233 1.20 2.34e- 1
5 NasalCongestionYes 0.0556 0.229 0.243 8.09e- 1
6 CoughYNYes -0.210 0.427 -0.491 6.24e- 1
7 SneezeYes -0.175 0.193 -0.906 3.66e- 1
8 FatigueYes -0.527 0.411 -1.28 2.02e- 1
9 SubjectiveFeverYes 0.266 0.195 1.37 1.74e- 1
10 HeadacheYes 0.128 0.263 0.487 6.27e- 1
# … with 28 more rowsbt_aug_test_all <- augment(bt_fit_test_all, test_data)Warning in predict.lm(object = object$fit, newdata = new_data, type =
"response"): prediction from a rank-deficient fit may be misleading### RMSE and R2 to test model performance
rmse2 <- bt_aug_test_all %>% rmse(truth = BodyTemp, .pred) #RMSE
rsq2 <- bt_aug_test_all %>% rsq(truth = BodyTemp, .pred) #RSQ
bt_metrics_all_test<- full_join(rmse2, rsq2)Joining with `by = join_by(.metric, .estimator, .estimate)`bt_metrics_all_test# A tibble: 2 × 3
.metric .estimator .estimate
<chr> <chr> <dbl>
1 rmse standard 0.950
2 rsq standard 0.223Here it looks like the test data outperformed the model data (lower rmse and higher rsq).
Model 4 fitting: runny nose to predict Body Temperature
Create new recipe and workflow
recipe_bt2 <- recipe(BodyTemp ~RunnyNose, data = train_data) #only include runny nose
bt_lin_wf2 <- workflow() %>%
add_model(lin_mod) %>% #model definition, use the same as Model 1
add_recipe(recipe_bt2) #model recipeModel fitting
bt_fit2 <- bt_lin_wf2 %>%
fit(data = train_data)
bt_fit2 %>% extract_fit_parsnip() %>%
tidy()# A tibble: 2 × 5
term estimate std.error statistic p.value
<chr> <dbl> <dbl> <dbl> <dbl>
1 (Intercept) 99.2 0.0947 1048. 0
2 RunnyNoseYes -0.318 0.114 -2.80 0.00537Model assessment on training data
set.seed(626)
bt_aug_train_uni <- augment(bt_fit2, train_data) #Train Data
### RMSE and R2 to test model performance
rmse3 <- bt_aug_train_uni %>% rmse(truth = BodyTemp, .pred) #RMSE
rsq3 <- bt_aug_train_uni %>% rsq(truth = BodyTemp, .pred) #RSQ
bt_metrics_uni_train<- full_join(rmse3, rsq3)Joining with `by = join_by(.metric, .estimator, .estimate)`bt_metrics_uni_train# A tibble: 2 × 3
.metric .estimator .estimate
<chr> <chr> <dbl>
1 rmse standard 1.22
2 rsq standard 0.0141Model assessment on testing data
set.seed(626)
bt_fit_test_uni <- bt_lin_wf2 %>%
fit(data = test_data)
bt_fit_test_uni %>% extract_fit_parsnip() %>%
tidy()# A tibble: 2 × 5
term estimate std.error statistic p.value
<chr> <dbl> <dbl> <dbl> <dbl>
1 (Intercept) 99.0 0.163 607. 2.12e-301
2 RunnyNoseYes -0.172 0.187 -0.918 3.60e- 1bt_aug_test_uni <- augment(bt_fit_test_uni, test_data) #Test Data
### RMSE and R2 to test model performance
rmse4 <- bt_aug_test_uni %>% rmse(truth = BodyTemp, .pred) #RMSE
rsq4 <- bt_aug_test_uni %>% rsq(truth = BodyTemp, .pred) #RSQ
bt_metrics_uni_test<- full_join(rmse4, rsq4)Joining with `by = join_by(.metric, .estimator, .estimate)`bt_metrics_uni_test# A tibble: 2 × 3
.metric .estimator .estimate
<chr> <chr> <dbl>
1 rmse standard 1.08
2 rsq standard 0.00464Here, the training data outperformed the model data, but either way, it does not look like Runny nose is a good predictor of Body Temperature.
Ultimately, our multivariate linear regression provided better performance assessments, so there is more exploring to be done!